7 Full workflow
7.1 Preface
This is an example of me (Julie) “talking” the reader through conducting an iSSA on the fisher data that is set up in our targets workflow.
7.2 Preparing an iSSA
7.2.1 The data
The critical data for running an iSSA are the x and y coordinates for GPS relocations and the associated datetime timestamp. Depending on what you’re doing, you’ll probably want to know the individual that those relocations belong to as well.
There are several things that need to be done prior to actually doing an iSSA
Overarchingly:
- Clean the GPS data
- Prepare the layers and other covariate data
7.2.1.1 Clean the GPS data
Things that you need to check and clean if they are not met:
- Make sure all observations (GPS points) are complete
- if not, remove incomplete observations
- Check if there are duplicated GPS points (two taken at the same time)
- remove duplicate observations
7.2.1.2 Prepare covariate input data
It is critical that all of the layers you plan to use and GPS points are in the same coordinate reference system. This can take a long time to reproject layers, crop them, and anything else you need to do for everything to be in the same “world,” but it is critical to do for the later steps of extracting covariates by step.
7.2.2 Making tracks
Once the data is clean, we make the points into a track. If you have more than one individual in the dataset, do it by individual.
Generating tracks:
make_track(locs_prep, x_, y_, t_, all_cols = TRUE, crs = epsg)7.2.2.1 Check the sampling rate
Now we should check how the sampling rate looks due to the messiness of GPS relocations.
summarize_sampling_rate_many(tar_read(tracks), 'id')| id | min | q1 | median | mean | q3 | max | sd | n | unit |
|---|---|---|---|---|---|---|---|---|---|
| 1072 | 3.92 | 9.8 | 10.0 | 20.7 | 10.3 | 1650 | 95 | 1348 | min |
| 1465 | 0.40 | 2.0 | 2.0 | 8.9 | 4.0 | 1080 | 48 | 3003 | min |
| 1466 | 0.42 | 2.0 | 2.1 | 15.7 | 4.1 | 2167 | 104 | 1500 | min |
| 1078 | 1.35 | 9.8 | 10.0 | 21.7 | 10.3 | 2111 | 87 | 1637 | min |
| 1469 | 0.42 | 2.0 | 2.2 | 13.2 | 5.4 | 2889 | 90 | 2435 | min |
| 1016 | 0.07 | 1.9 | 2.0 | 8.0 | 2.5 | 1209 | 44 | 8957 | min |
Data are messy. For iSSA, we need the GPS fixes to happen at regular intervals, the same for all individuals in the model, so we likely need to resample the data.
resample_tracks(tracks, rate = rate, tolerance = tolerance)In our targets workflow, we have simplified all of this. Here, all you will
have to do is at the beginning of the workflow: fill in
the Variables to match your data.
# Variables ---------------------------------------------------------------
# Targets: prepare
id_col <- 'id'
datetime_col <- 't_'
x_col <- 'x_'
y_col <- 'y_'
epsg <- 32618
crs <- st_crs(epsg)
crs_sp <- CRS(crs$wkt)
tz <- 'America/New_York'
# Targets: tracks
# Split by: within which column or set of columns (eg. c(id, yr))
# do we want to split our analysis?
split_by <- id_col
# Resampling rate
rate <- minutes(30)
# Tolerance
tolerance <- minutes(5)
# Number of random steps
n_random_steps <- 107.2.3 Steps
Now we have tracks, but a step selection analysis requires steps! So, in our targets workflow, we only keep data for individuals that have at least 3 steps, and this is adjustable depending on what you need.
7.2.3.1 Check the distribution of steps and turn angles
Now to make sure there aren’t any biological impossibilities in the steps.
Usually there are a lot of little steps and few big steps, but this is a good
check to make sure any erroneous observations are removed. For instance, this
would show if there are any steps that are not biologically possible. It also
shows the distribution so you know if you should use a Gamma or exponential
distribution for generating random steps. Most are Gamma, and that is the
default in amt.
SL distribution for 1 individual
tar_read(dist_sl_plots, 1)## $dist_sl_plots_8ba21cb0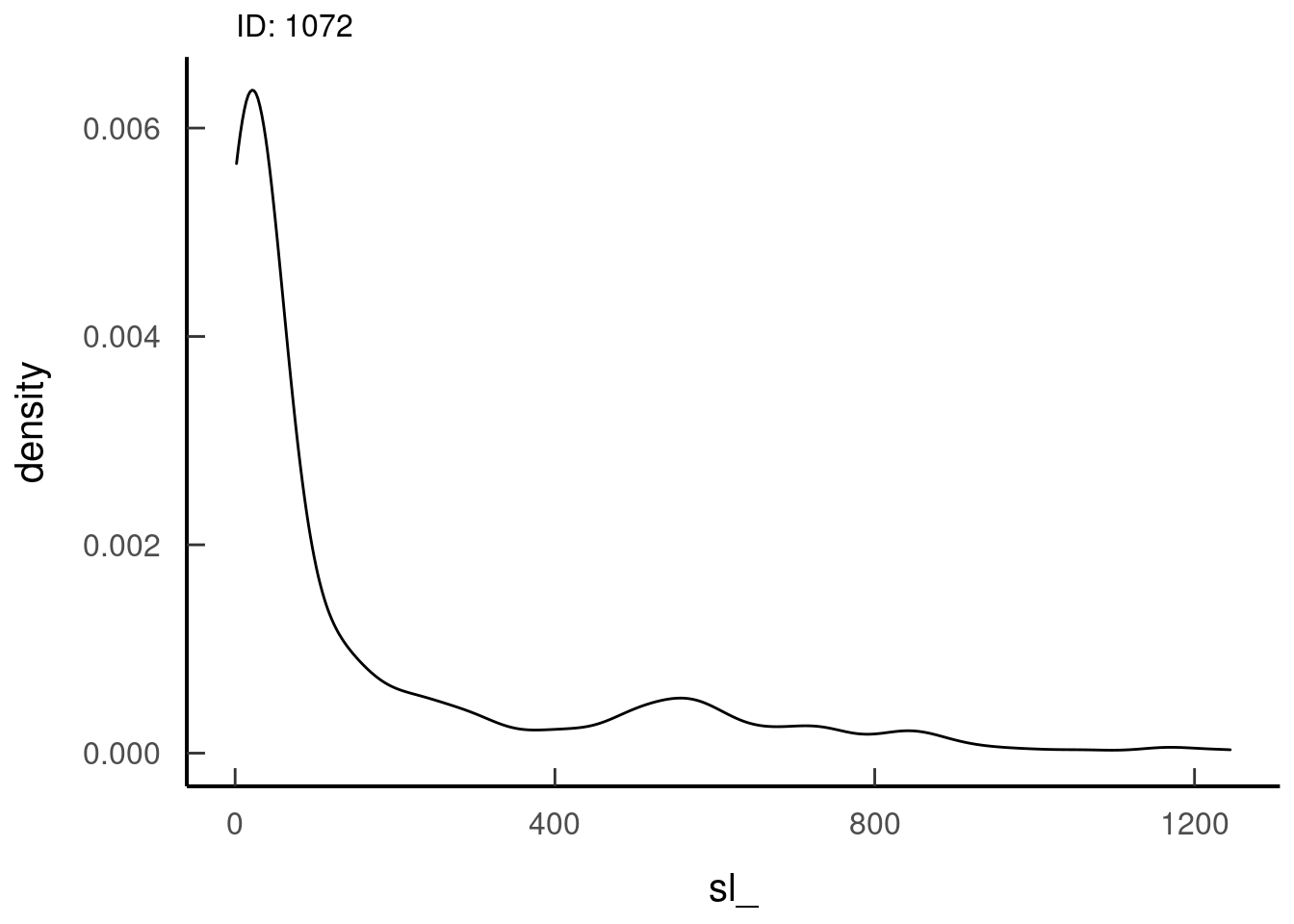 Looks like a gamma distribution, so that’s good
Looks like a gamma distribution, so that’s goodTA distribution for 1 individual
tar_read(dist_ta_plots, 1)## $dist_ta_plots_8ba21cb0## Warning: Removed 35 rows containing non-finite values (stat_density).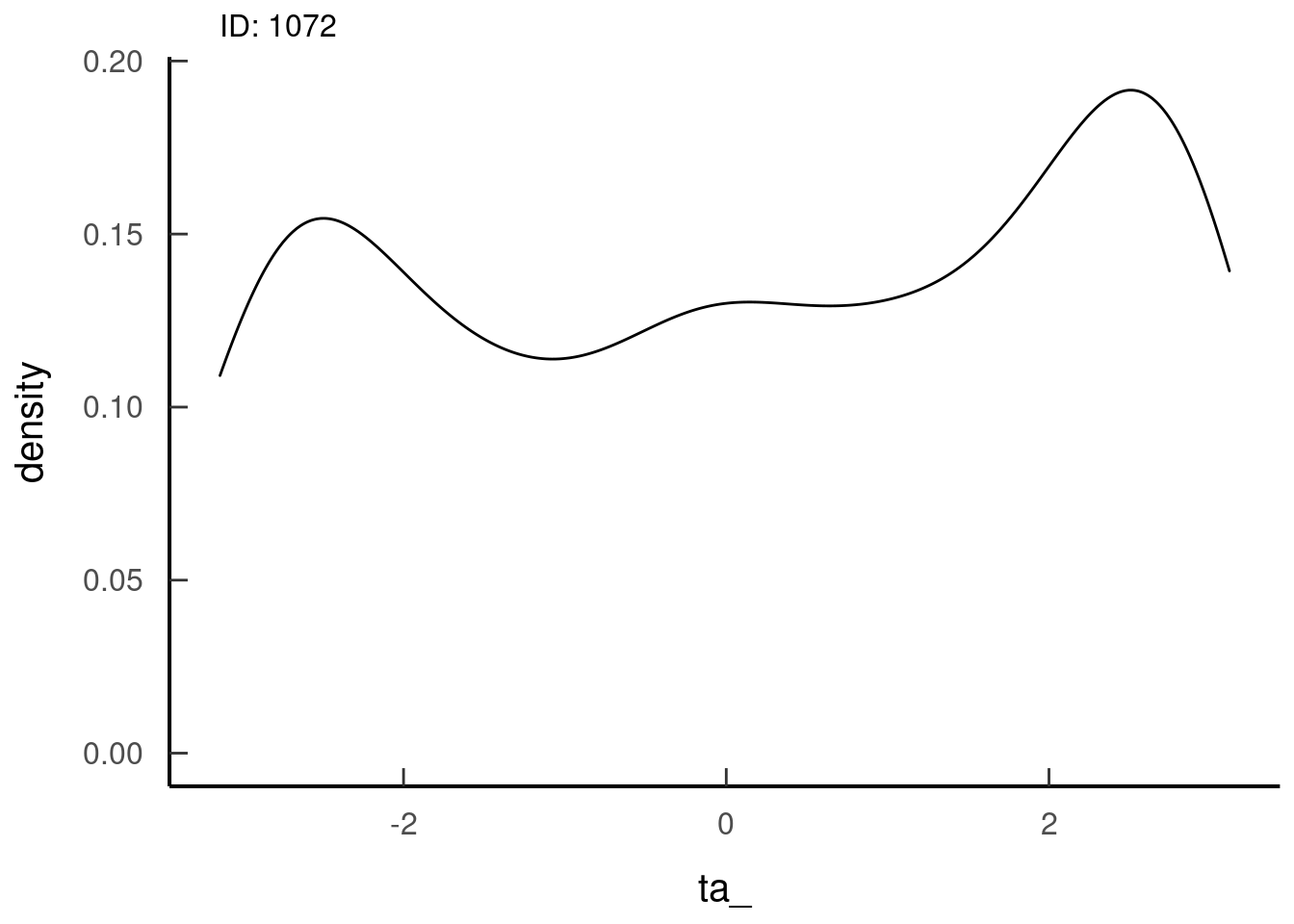 Looks like a von Mises distribution, so that’s also good
Looks like a von Mises distribution, so that’s also good7.2.4 Prepare the data to go into the iSSA
So, at this point, we have our observed steps and know what their distributions look like. Now we can use this information to properly inform our random steps with the correct distribution. As we create the random steps, we will also extract the covariates of interest at each step.
7.2.4.1 Motivation vs Selection
One of the critical decisions to make in your analysis is if you want to know if a variable is motivating why/how an animal moves vs. if the animal is selecting for that variable; this decision influences if you use the information from the start of the step (motivation) vs. the end of the step (selection) in your analysis. This decision is of course determined by your question and hypotheses
See Choosing covariates for more info
7.2.4.2 Example extraction
For example, here we are creating random steps and extracting information about what landcover class and the distance from water an individual selects (at the end of each step) and time of day (day or night at the start of the step) at the time of each step. This will create a dataset that includes step length, turn angle, whether it’s day or night at the start of the step, and what land class an animal is on at the end of their step.
NOTE: In the targets workflow, we have created out own extraction functions
instead of using amt’s extract_covariate and time_of_day functions. This
is for several reasons - more flexibility - amt isn’t functional with terra
yet (coming soon) - time_of_day is good for day vs night, but if you want dawn
or dusk, it’s not great at larger fixrates. For instance, if you have a 2 hour
fixrate, it rarely includes dawn or dusk because those time frames are narrow in
the default maptools function that time_of_day wraps around. In this
circumstance, we have created our own time of day extractions as needed by
location. However, for our fisher example here with a 30 minute fixrate,
including default crepuscular times should be fine.
# R/extract_layers.R
extract_layersfunction (DT, crs, lc, legend, elev, popdens, water)
{
setDT(DT)
start <- c("x1_", "y1_")
end <- c("x2_", "y2_")
extract_pt(DT, lc, end)
DT[legend, `:=`(lc_description, description), on = .(pt_lc = Value)]
extract_pt(DT, elev, end)
extract_pt(DT, popdens, end)
extract_distance_to(DT, water, end, crs)
}If you have multiple hypotheses that would benefit from know information from
both the beginning and the end of the step, you can input"both" instead of
"start" or "end".
So, in the end, we get a dataset that looks something like:
| x1_ | x2_ | y1_ | y2_ | sl_ | ta_ | id | tar_group | burst_ | t1_ | t2_ | dt_ | step_id_ | case_ | pt_lc | lc_description | pt_elev | pt_popdens | dist_to_water |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 593975 | 593969 | 4739337 | 4739492 | 155.61 | NA | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | TRUE | 41 | forest | 83 | 51 | 171 |
| 593975 | 593967 | 4739337 | 4739367 | 31.62 | 0.21 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 42 | forest | 100 | 102 | 289 |
| 593975 | 593959 | 4739337 | 4739362 | 29.55 | 0.52 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 42 | forest | 100 | 102 | 297 |
| 593975 | 594216 | 4739337 | 4739595 | 353.46 | -0.79 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 21 | developed | 58 | 51 | 49 |
| 593975 | 593973 | 4739337 | 4739338 | 2.03 | 0.90 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 42 | forest | 105 | 102 | 314 |
| 593975 | 594090 | 4739337 | 4739452 | 162.43 | -0.83 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 41 | forest | 77 | 51 | 180 |
| 593975 | 593978 | 4739337 | 4739339 | 4.15 | -0.93 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 42 | forest | 105 | 102 | 311 |
| 593975 | 593829 | 4739337 | 4739372 | 150.09 | 1.29 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 43 | forest | 90 | 102 | 333 |
| 593975 | 593916 | 4739337 | 4739366 | 66.11 | 1.06 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 43 | forest | 97 | 102 | 305 |
| 593975 | 593975 | 4739337 | 4739337 | 0.48 | 1.41 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 42 | forest | 105 | 102 | 314 |
| 593975 | 594066 | 4739337 | 4739605 | 283.09 | -0.37 | 1072 | 1 | 4 | 2011-02-13 04:31:32 | 2011-02-13 05:00:13 | 29 mins | 1 | FALSE | 41 | forest | 72 | 51 | 32 |
7.3 Running an iSSA
This workflow is going to be an example using the Poisson point process method described by Muff et al (2020). This method allows us to look at all individuals at once so we can examine both the individual- and population-level responses at once. In this example, we’re going to include both categorical and continuous variables.
NOTE: Random effects in GLMMs are supposed to have 5 or more levels, which means you should have at least 5 individuals if you want to include them as a random effect.
# R/model_land_cover.R
model_land_coverfunction (DT)
{
glmmTMB(case_ ~ -1 + I(log(sl_)) + I(log(sl_)):lc_adj + lc_adj +
I(log(dist_to_water + 1)) + I(log(dist_to_water + 1)):I(log(sl_)) +
(1 | indiv_step_id) + (0 + I(log(sl_)) | id) + (0 + I(log(sl_)):lc_adj |
id) + (0 + lc_adj | id) + (0 + I(log(dist_to_water +
1)) | id) + (0 + I(log(dist_to_water + 1)):I(log(sl_)) |
id), data = DT, family = poisson(), map = list(theta = factor(c(NA,
1:23))), start = list(theta = c(log(1000), seq(0, 0,
length.out = 23))))
}
summary(tar_read(model_lc))## Family: poisson ( log )
## Formula:
## case_ ~ -1 + I(log(sl_)) + I(log(sl_)):lc_adj + lc_adj + I(log(dist_to_water +
## 1)) + I(log(dist_to_water + 1)):I(log(sl_)) + (1 | indiv_step_id) +
## (0 + I(log(sl_)) | id) + (0 + I(log(sl_)):lc_adj | id) +
## (0 + lc_adj | id) + (0 + I(log(dist_to_water + 1)) | id) +
## (0 + I(log(dist_to_water + 1)):I(log(sl_)) | id)
## Data: DT
##
## AIC BIC logLik deviance df.resid
## NA NA NA NA 26488
##
## Random effects:
##
## Conditional model:
## Groups Name Variance Std.Dev. Corr
## indiv_step_id (Intercept) 1.00e+06 1.00e+03
## id I(log(sl_)) 1.87e-90 1.37e-45
## id.1 I(log(sl_)):lc_adjdisturbed 1.08e-02 1.04e-01
## I(log(sl_)):lc_adjforest 2.05e-03 4.52e-02 -1.00
## I(log(sl_)):lc_adjopen 1.08e-01 3.28e-01 1.00
## I(log(sl_)):lc_adjwetlands 2.05e-03 4.53e-02 -1.00
## id.2 lc_adjdisturbed 4.42e-01 6.64e-01
## lc_adjforest 3.32e-02 1.82e-01 -1.00
## lc_adjopen 4.42e+00 2.10e+00 1.00
## lc_adjwetlands 2.38e-26 1.54e-13 -0.27
## id.3 I(log(dist_to_water + 1)) 1.34e-02 1.16e-01
## id.4 I(log(dist_to_water + 1)):I(log(sl_)) 5.26e-90 2.29e-45
##
##
##
##
##
## -1.00
## 1.00 -1.00
##
##
## -1.00
## 0.30 -0.28
##
##
## Number of obs: 26521, groups: indiv_step_id, 2411; id, 6
##
## Conditional model:
## Estimate Std. Error z value Pr(>|z|)
## I(log(sl_)) -0.13107 NaN NaN NaN
## lc_adjdisturbed -3.74565 NaN NaN NaN
## lc_adjforest -3.42646 NaN NaN NaN
## lc_adjopen -5.94910 NaN NaN NaN
## lc_adjwetlands -3.67823 NaN NaN NaN
## I(log(dist_to_water + 1)) -0.03581 NaN NaN NaN
## I(log(sl_)):lc_adjforest 0.23726 NaN NaN NaN
## I(log(sl_)):lc_adjopen 0.29305 NaN NaN NaN
## I(log(sl_)):lc_adjwetlands 0.28385 NaN NaN NaN
## I(log(sl_)):I(log(dist_to_water + 1)) 0.00072 NaN NaN NaNWhen looking at the output of this model, we see some issues. First of all,
there are some warnings from the model. The positive hessian warning is applied
liberally in glmmTMB, but the real sign of a problem is the NAs reported for
variation. This model didn’t converge properly, so we need to look at why.
Things to explore at the data level include:
- lots of
NAsfor a particular variable (helpful tips from Bolker) - lots of variation or very little variation in a particular variable
- availability of variables for individuals
Things to explore at the model level include:
- the model is over-parameterized for the amount of data
- the high variance is assigned at the strata level and not for another random effect
- variables are similarly scaled
- if using the Muff method, the strata represent the step id by individual
(
indiv_step_id), not the step id that is assigned byamtbecause the way we have it coded, the numbers restart for each individual - categorical variables are classified as factors, especially
indiv_step_id - random effects have at least 5 levels (this is likely not going to be a problem for most iSSAs, but it has come up)
# checking availability
# for landcover
tar_read(avail_lc)| id | prop_lc_description | lc_description |
|---|---|---|
| 1072 | 0.72 | forest |
| 1072 | 0.16 | developed |
| 1072 | 0.01 | water |
| 1072 | 0.02 | cultivated |
| 1072 | 0.07 | wetlands |
| 1072 | 0.00 | barren |
| 1072 | 0.00 | shrubland |
| 1465 | 0.37 | wetlands |
| 1465 | 0.19 | developed |
| 1465 | 0.37 | forest |
| 1465 | 0.07 | cultivated |
| 1465 | 0.01 | barren |
| 1465 | 0.00 | water |
| 1465 | 0.00 | shrubland |
| 1466 | 0.61 | wetlands |
| 1466 | 0.17 | developed |
| 1466 | 0.05 | cultivated |
| 1466 | 0.15 | forest |
| 1466 | 0.01 | water |
| 1078 | 0.48 | forest |
| 1078 | 0.28 | developed |
| 1078 | 0.21 | wetlands |
| 1078 | 0.00 | shrubland |
| 1078 | 0.03 | cultivated |
| 1078 | 0.00 | water |
| 1469 | 0.18 | forest |
| 1469 | 0.06 | cultivated |
| 1469 | 0.42 | developed |
| 1469 | 0.33 | wetlands |
| 1469 | 0.01 | water |
| 1016 | 0.49 | forest |
| 1016 | 0.16 | developed |
| 1016 | 0.01 | shrubland |
| 1016 | 0.20 | wetlands |
| 1016 | 0.06 | cultivated |
| 1016 | 0.08 | water |
| 1016 | 0.00 | herbaceous |
# for distance to water
tracks_extract <- tar_read(tracks_extract)
tracks_extract[!(case_), quantile(dist_to_water, na.rm = TRUE), by = id]| id | V1 |
|---|---|
| 1072 | 0 |
| 1072 | 285 |
| 1072 | 1131 |
| 1072 | 1714 |
| 1072 | 3049 |
| 1465 | 0 |
| 1465 | 595 |
| 1465 | 854 |
| 1465 | 1188 |
| 1465 | 2573 |
| 1466 | 0 |
| 1466 | 222 |
| 1466 | 418 |
| 1466 | 809 |
| 1466 | 2154 |
| 1078 | 0 |
| 1078 | 1833 |
| 1078 | 2271 |
| 1078 | 2604 |
| 1078 | 3376 |
| 1469 | 0 |
| 1469 | 418 |
| 1469 | 791 |
| 1469 | 1365 |
| 1469 | 2594 |
| 1016 | 0 |
| 1016 | 135 |
| 1016 | 442 |
| 1016 | 918 |
| 1016 | 1839 |
After some exploration, I’m thinking this model is over-parameterized for 6 individuals, at least having so many landcover classes. So, I’m going to pretend for this example that we only care about selection for forest and disturbed habitats. We created dummy variables for this based on the extracted landcover data and ran the iSSA on this data.
# R/model_forest_bin.R
model_forest_binfunction (DT)
{
glmmTMB(case_ ~ -1 + I(log(sl_)) + I(log(sl_)):forest + forest +
I(log(sl_)):disturbed + disturbed + I(log(dist_to_water +
1)) + I(log(dist_to_water + 1)):I(log(sl_)) + (1 | indiv_step_id) +
(0 + I(log(sl_)) | id) + (0 + I(log(sl_)):disturbed |
id) + (0 + I(log(sl_)):forest | id) + (0 + forest | id) +
(0 + disturbed | id) + (0 + I(log(dist_to_water + 1)) |
id) + (0 + I(log(dist_to_water + 1)):I(log(sl_)) | id),
data = DT, family = poisson(), map = list(theta = factor(c(NA,
1:7))), start = list(theta = c(log(1000), seq(0,
0, length.out = 7))))
}
summary(tar_read(model_forest))## Family: poisson ( log )
## Formula:
## case_ ~ -1 + I(log(sl_)) + I(log(sl_)):forest + forest + I(log(sl_)):disturbed +
## disturbed + I(log(dist_to_water + 1)) + I(log(dist_to_water +
## 1)):I(log(sl_)) + (1 | indiv_step_id) + (0 + I(log(sl_)) |
## id) + (0 + I(log(sl_)):disturbed | id) + (0 + I(log(sl_)):forest |
## id) + (0 + forest | id) + (0 + disturbed | id) + (0 + I(log(dist_to_water +
## 1)) | id) + (0 + I(log(dist_to_water + 1)):I(log(sl_)) | id)
## Data: DT
##
## AIC BIC logLik deviance df.resid
## 49162 49276 -24567 49134 26507
##
## Random effects:
##
## Conditional model:
## Groups Name Variance Std.Dev.
## indiv_step_id (Intercept) 1.00e+06 1.00e+03
## id I(log(sl_)) 1.26e-09 3.55e-05
## id.1 I(log(sl_)):disturbed 7.67e-04 2.77e-02
## id.2 I(log(sl_)):forest 2.65e-10 1.63e-05
## id.3 forest 1.77e-02 1.33e-01
## id.4 disturbed 1.23e-10 1.11e-05
## id.5 I(log(dist_to_water + 1)) 1.34e-02 1.16e-01
## id.6 I(log(dist_to_water + 1)):I(log(sl_)) 1.55e-11 3.93e-06
## Number of obs: 26521, groups: indiv_step_id, 2411; id, 6
##
## Conditional model:
## Estimate Std. Error z value Pr(>|z|)
## I(log(sl_)) 0.07923 0.07735 1.02 0.306
## forest 0.34653 0.19410 1.79 0.074 .
## disturbed -0.19495 0.27753 -0.70 0.482
## I(log(dist_to_water + 1)) 0.08271 0.08956 0.92 0.356
## I(log(sl_)):forest -0.05632 0.03635 -1.55 0.121
## I(log(sl_)):disturbed -0.24794 0.05330 -4.65 3.3e-06 ***
## I(log(sl_)):I(log(dist_to_water + 1)) 0.00875 0.01142 0.77 0.444
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1This model runs without issue.
7.4 Interpretation
Demonstrating effects from iSSA output is an area where you will see a lot of different avenues at this point. Some just show the beta estimates from the model and p-values. This, however can result in misinterpretation of biological significance (Avgar et al. 2017).
7.4.1 Relative Selection Strength
This is where Relative Selection Strength (RSS) comes in. Avgar et al (2017)
derived various equations to calculate RSS depending on if variables are on
their own, interacted with others, log-transformed, etc. However, since then
Brian Smith has determined that using the predict() function can achieve the
same outcome (see proof Smith 2020).
But this is all the mathematical details.
What RSSs are really doing is comparing the selection strength for habitat at location 1 (H1) compared to location 2 (H2).So, say an animal is not in a forest, are they relatively
more likely to select to be in a forest or avoid the forest? Then the RSS shows
the strength of selection or avoidance over a range of habitat values of that
habitat type.
In the targets-issa workflow, we predict H1, and H2 for forest using the following functions:
# R/predict_h1_forest.R
predict_h1_forest## function (DT, model)
## {
## N <- 100L
## new_data <- DT[, .(sl_ = mean(sl_), forest = seq(from = 0,
## to = 1, length.out = N), disturbed = 0, dist_to_water = median(dist_to_water,
## na.rm = TRUE), indiv_step_id = NA), by = id]
## new_data[, `:=`(h1_forest, predict(model, .SD, type = "link",
## re.form = NULL))]
## new_data[, `:=`(x, seq(from = 0, to = 1, length.out = N)),
## by = id]
## }
# R/predict_h2.R
predict_h2## function (DT, model)
## {
## new_data <- DT[, .(sl_ = mean(sl_), forest = 0, disturbed = 0,
## dist_to_water = median(dist_to_water, na.rm = TRUE),
## indiv_step_id = NA), by = id]
## new_data[, `:=`(h2, predict(model, .SD, type = "link", re.form = NULL))]
## }
# Then we can calculate RSS for forest as H1 - H2:
# R/calc_rss.R
calc_rss## function (pred_h1, h1_col, pred_h2, h2_col)
## {
## log_rss <- merge(pred_h1[, .SD, .SDcols = c("id", "x", h1_col)],
## pred_h2[, .SD, .SDcols = c("id", h2_col)], by = "id",
## all.x = TRUE)
## log_rss[, `:=`(rss, h1 - h2), env = list(h1 = h1_col, h2 = h2_col)]
## }And finally, the plots with the plot_rss function.
tar_read(plot_rss_forest)## `geom_smooth()` using method = 'loess' and formula 'y ~ x'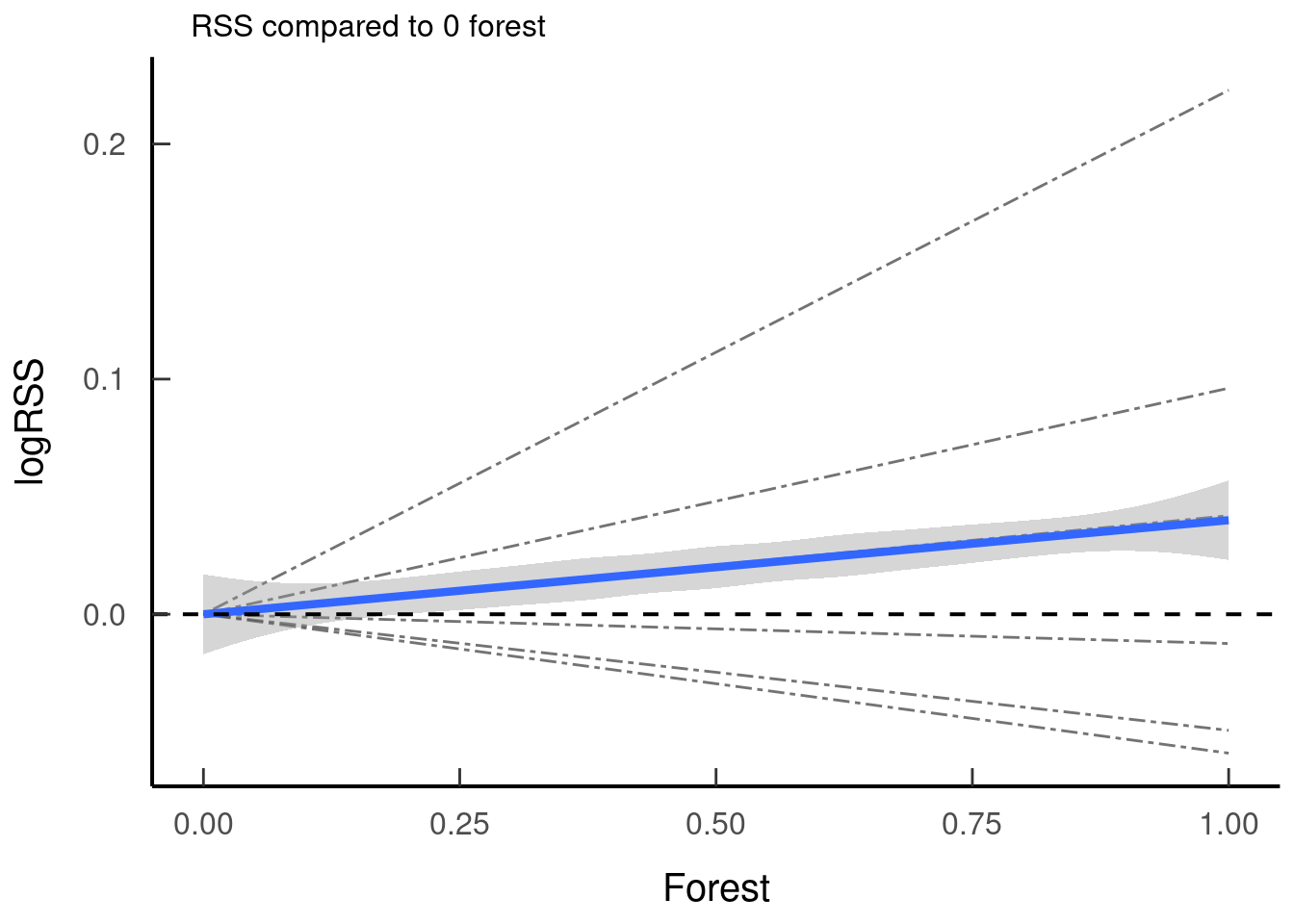
JWT comment: I personally find it hard to wrap my head around RSSs for categorical variables, and I don’t have a great way of explaining it for others like me who didn’t get it. The closest I can describe is, say, looking at this plot, the individual starts not in forest, is their next step more likely to go to forest or not. To continuous nature of the RSS is hard for me to wrap my head around with a categorical variable. I just pretend in my head that it’s a proportional value of ‘no forest’ = 0 and ‘all forest’ = 1, and the middle is an estimate based on the model. It’s not perfect, but that’s the best I’ve been able to come to terms with it. :shrug: #OverlyHonestMethods
We can then do the same thing to determine the RSS for distance to water.
tar_read(plot_rss_water)## `geom_smooth()` using method = 'loess' and formula 'y ~ x'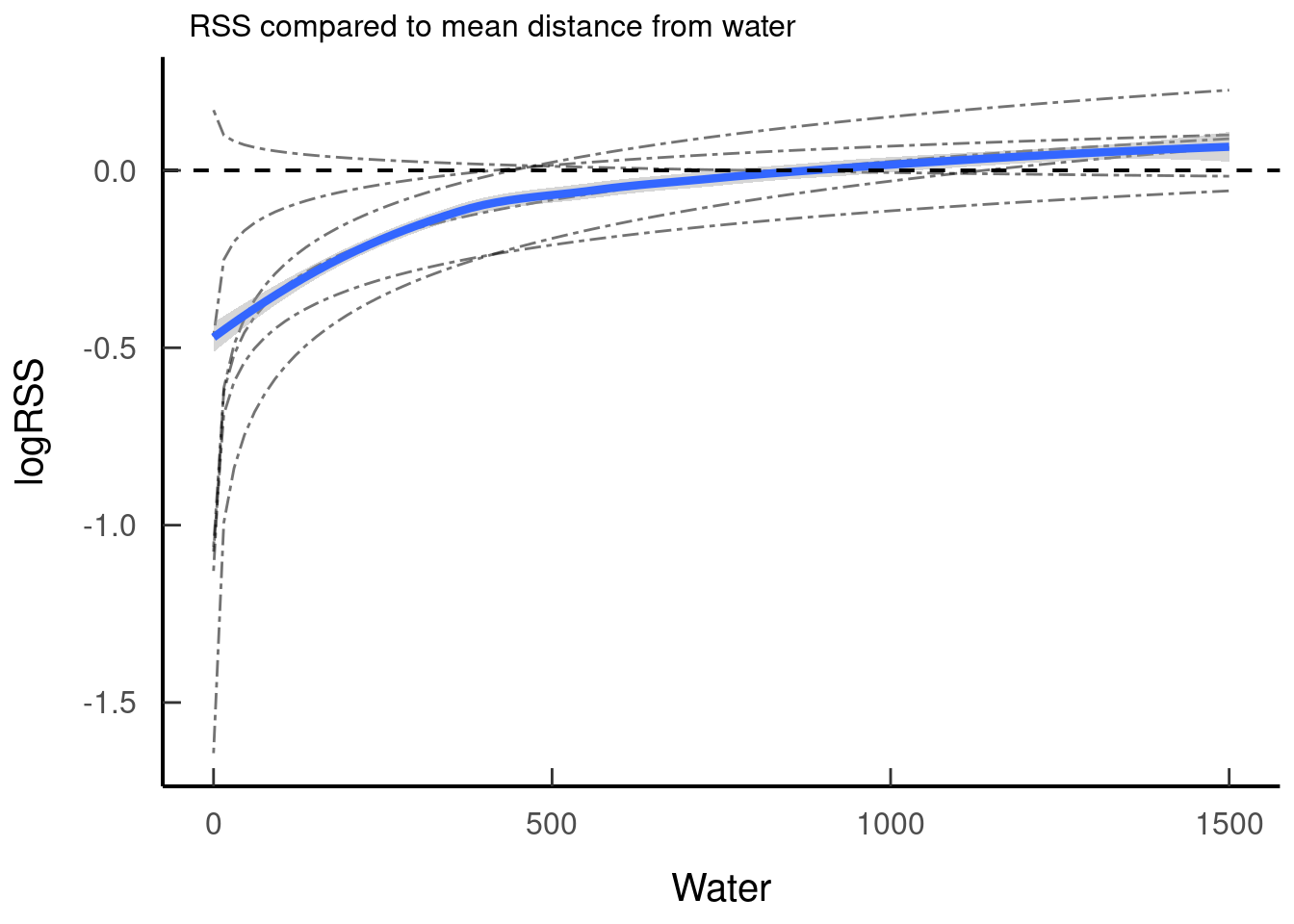
RSSs have to be done one habitat at a time. That being said, interactions can be explored by pre-determining cut off points to examine one of the variables at a time. I know that sounds confusing. So, for example, we have selection of forest interacted with distance to road. Say we want to understand how animals select for forest when they are near or far from roads. So, we define being near a road as being 100m of the road and being far from the road as 5000m from the road. These values are semi-arbitrary, but they should be based on the biology of the animal. This complicates the calculation of the RSS a bit, making it a two step process with the predict method. You first have to calculate the overall selection for, in this case, forest at 100m from the road, then you run the run the predict function over the availability of forest and add them together.
7.4.2 Speed
Speed is a different kettle of fish. Some people show box plots of step length to give an idea of speed, but this is only a partial show of effect size (this is not phrased well – fix). When using a gamma distribution for an iSSA, the mean estimated speed is shape parameter multiplied by the scale parameter from the gamma distribution. However, the shape is modified by the movement variables in the iSSA itself. So this calculation becomes
\(\text{mean estimated speed} = (shape + logSL.beta + logSLintx.beta*mean(what.interacted.with))*scale\)
So, from our toy example, this could be done as below. Here, we can pick which
covariate we want to vary to estimate speed to see how speed is impacted by what
it is interacted with. seq represents the range of values we’re looking over.
# R/calc_speed.R
calc_speed## function (DT, covariate, seq)
## {
## if (covariate == "forest")
## DT[, `:=`(spd = list(list((shape + `I(log(sl_))` + `I(log(sl_)):forest` *
## seq + `I(log(sl_)):disturbed` + `I(log(dist_to_water + 1)):I(log(sl_))` *
## median.water) * (scale))), x = list(list(seq))),
## by = .(id)]
## if (covariate == "disturbed")
## DT[, `:=`(spd = list(list((shape + `I(log(sl_))` + `I(log(sl_)):forest` +
## `I(log(sl_)):disturbed` * seq + `I(log(dist_to_water + 1)):I(log(sl_))` *
## median.water) * (scale))), x = list(list(seq))),
## by = .(id)]
## if (covariate == "dist_to_water")
## DT[, `:=`(spd = list(list((shape + `I(log(sl_))` + `I(log(sl_)):forest` +
## `I(log(sl_)):disturbed` + `I(log(dist_to_water + 1)):I(log(sl_))` *
## seq) * (scale))), x = list(list(seq))), by = .(id)]
## move <- DT[, .(spd = unlist(spd), x = unlist(x)), by = .(id)]
## move
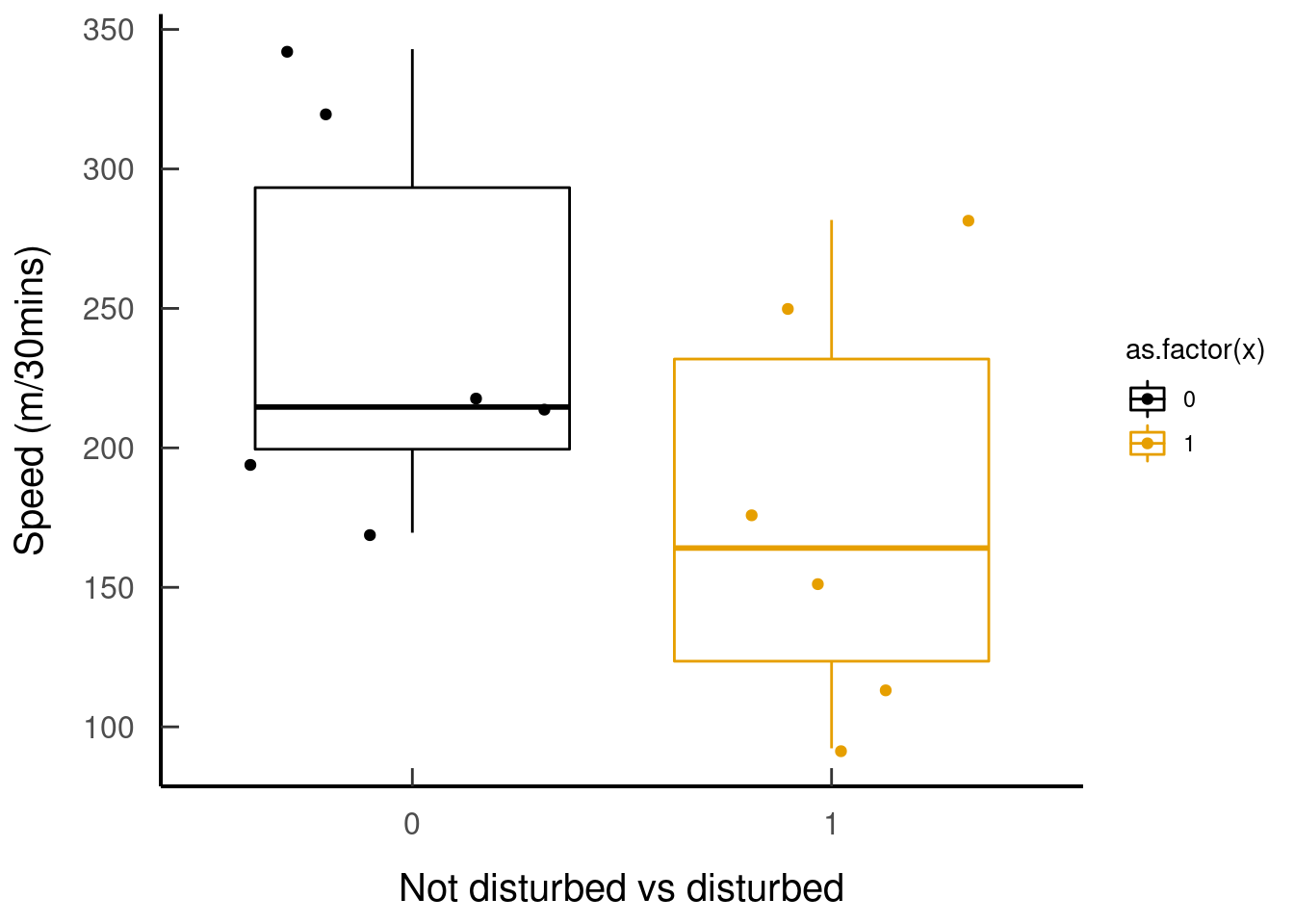
## }Let’s see how fishers’ speed changes in areas without anthropogenic disturbance vs areas that are disturbed (seq = 0:1)
tar_read(plot_speed_disturbed)